Translate this page into:
Sterile body fluids infections: Profile of bacteria and their antimicrobial resistance pattern in a tertiary care hospital from Uttar Pradesh
*Corresponding author: Peetam Singh, Department of Microbiology, Subharti Medical College, Meerut, Uttar Pradesh, India. kgmclko@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Singh P, Pandey A, Bisht AS. Sterile body fluids infections: Profile of bacteria and their antimicrobial resistance pattern in a tertiary care hospital from Uttar Pradesh. Indian J Med Sci 2023;75:161-7.
Abstract
Objectives:
Infections of sterile body fluids are important and significant causes of mortality and morbidity, especially healthcare-associated infections. Species-level identification and antimicrobial resistance profile of bacteria are important determinants while selecting appropriate antimicrobials for empirical and targeted therapy. We conducted this study to observe the distribution of various bacteria and their antimicrobial resistance profile isolated from sterile body fluids.
Materials and Methods:
We conducted this study in a tertiary care teaching hospital from western Uttar Pradesh for a period of 2 years. All sterile body fluid samples were processed by conventional aerobic bacterial culture followed by their identification up to species level by conventional biochemicals following standard microbiological procedures. The antimicrobial susceptibility of the bacterial pathogens grown in culture was tested by Kirby–Bauer disk diffusion method and interpretation of susceptibility testing was done according to CLSI guidelines 2020.
Results:
A total of 1980 sterile body fluid samples were collected during the study period and 192 samples were found positive on culture for bacterial pathogens. Gram-negative bacilli (GNB) were predominantly isolated, comprising 83.33% in comparison to 16.67 % of Gram-positive cocci. Among Staphylococcus aureus isolates, 75% were methicillin-resistant S. aureus. All S. aureus isolates were sensitive against vancomycin and linezolid. Among GNB, 25% were extended-spectrum beta-lactamase producers while 62.5% were carbapenemase producers. All GNBs were sensitive to colistin.
Conclusion:
From this study, we concluded that the pathogenic bacteria implicated in infections of sterile body fluids are predominantly multidrug-resistant. There is a huge variation in data on the distribution of bacterial species isolated from sterile body fluids and their antimicrobial resistance patterns from different geographical locations and healthcare settings. Thus, data from a particular healthcare setting are important for empirical treatment in that healthcare setting.
Keywords
Sterile body fluid infection
Antimicrobial resistance
Methicillin-resistant Staphylococcus aureus
Extended-spectrum beta-lactamase
INTRODUCTION
The various body cavities are sterile sites and these body cavities are filled with sterile body fluids. The purpose of these body fluids is to bathe the organs and membranes for the protection of vital organs, transportation of nutrients, regulation of body temperature, and reducing friction.[1,2] The body fluids are generally sterile; however, different types of microorganisms can invade the body cavities leading to infection and physicochemical changes in these body fluids. Early identification of the organisms causing sterile body fluid infection along with antimicrobial susceptibility can help clinicians to initiate early and targeted antimicrobial therapy.[3] The data on bacterial profile and their antimicrobial susceptibility pattern isolated from sterile body fluids can help to develop a local antibiogram of a particular healthcare setting. The knowledge of commonly isolated organisms from various sterile body fluids along with their antimicrobial susceptibility testing (AST) pattern in the form of antibiogram can help in the selection of appropriate empirical antibiotics.[4]
The culture positivity rate in sterile body fluid infections is comparatively low varying from 10% to 30%.[4] Moreover, the patients empirically treated with antibiotics before sample collection hinder the recovery of pathogenic bacteria on culture. However, multidrug-resistant organisms’ (MDROs) emergence is also becoming an important challenge for treating clinicians and there is an urgent need for judicious use of antibiotics which can significantly reduce morbidity, hospital stay, and mortality, ultimately leading to reduced cost of treatment among patients with these infections.[5]
Rapidly increasing multidrug resistance (MDR) against commonly used antimicrobials is an important public health issue of concern worldwide. The distribution profile of bacteria isolated from sterile body fluids and their drug resistance patterns needs to be collected from various tertiary care institutions located in various regions which can further guide us in the implementation of antimicrobial stewardship program nationally.
Therefore, we planned this study to know the distribution profile of various bacterial isolates implicated in sterile body fluid infections, along with their AST patterns which can help us while selecting appropriate empirical antimicrobial therapy at our institution for better patient care.
MATERIALS AND METHODS
This prospective hospital-based study was done at a tertiary care medical teaching hospital in western Uttar Pradesh, for a period of 2 years. The clinical sterile body fluid specimens from patients were collected following exclusion and inclusion criteria.
Inclusion criteria
The following criteria were included in the study:
All sterile body fluid samples
Patients of all age groups
Patients of all genders.
Exclusion criteria
The following criteria were excluded from the study:
Patients with a prior history of antibiotic therapy before sample collection
Samples other than sterile body fluids
Blood samples were excluded from the study.
Sample processing
Collected clinical samples of various sterile body fluids were processed by standard microbiological procedures including:
Direct microscopy of Gram-stained smear from the specimens
Culture: Samples were subjected to aerobic bacterial culture by inoculating onto chocolate agar, blood agar, and MacConkey agar plates. The clinical specimens were also subjected to enrichment by inoculating into brain heart infusion (BHI) broth. The inoculated agar plates and broth were placed in an aerobic incubator at 37ºC for 48 h. Culture plates and BHI broth were examined initially after 24 h and finally after 48 h for the appearance of any growth. The bacterial pathogens grown in culture media were identified using standard microbiological procedures and conventional biochemical tests.
AST: The isolated bacterial pathogens were subjected to AST performed by Kirby–Bauer disk diffusion method and interpretation as per CLSI 2020 guidelines.[6] The panel of antibiotics along with their disk content for Gram-positive cocci (GPC) and Gram-negative bacilli (GNB) is depicted in [Tables 1 and 2], respectively.
| Antibiotic | Strength |
|---|---|
| Penicillin-G | 10 µg |
| Ampicillin | 10 µg |
| Cefoxitin | 30 µg |
| Cotrimoxazole | 1.25/232.75 µg |
| Tetracycline | 30 µg |
| Erythromycin | 15 µg |
| Clindamycin | 2 µg |
| Moxifloxacin | 5 µg |
| Chloramphenicol | 30 µg |
| Gentamicin | 10 µg |
| Ofloxacin | 5 µg |
| Doxycycline | 30 µg |
| Teicoplanin | 30 µg |
| Linezolid | 30 µg |
| Vancomycin | 30 µg |
| High-level gentamicin | 120 µg |
| High-level streptomycin | 300 µg |
| Antibiotics | Strength |
|---|---|
| Ampicillin | 10 µg |
| Piperacillin | 100 µg |
| Piperacillin-Tazobactam | 100/10 µg |
| Amoxicillin-Clavulanic acid | 20/10 µg |
| Ampicillin-Sulbactam | 10/10 µg |
| Cotrimoxazole | 1.25/232.75 µg |
| Tetracycline | 30 µg |
| Chloramphenicol | 30 µg |
| Gentamicin | 10 µg |
| Ciprofloxacin | 5 µg |
| Cefixime | 5 µg |
| Ceftazidime | 30 µg |
| Ceftriaxone | 30 µg |
| Aztreonam | 30 µg |
| Cefepime | 30 µg |
| Amikacin | 30 µg |
| Tobramycin | 10 µg |
| Ertapenem | 10 µg |
| Meropenem | 10 µg |
| Imipenem | 30 µg |
| Colistin | 10 µg |
| Cefotaxime-clavulanic acid | 30/10 µg |
| Cefotaxime | 30 µg |
| Ceftazidime-clavulanic acid | 30/10 µg |
AST of colistin against GNB was performed using Minimum inhibitory concentration (MIC) by broth microdilution and AST of vancomycin against Staphylococci was performed using MIC by E test.
Among GNB, extended-spectrum beta-lactamase (ESBL) was detected by phenotypic methods using cephalosporin/ clavulanate combination discs (cefotaxime 30 µg and ceftazidime 30 µg with and without clavulanate 10 µg). The modified Hodge test was used for phenotypic detection of carbapenemase production.
RESULTS
During the 2 years of the study period, 1980 sterile body fluid samples collected for culture and AST were processed. The pathogenic bacteria grown in 192 samples and culture positivity rate were found to be 9.7%. The culture positivity rate of various sterile body fluid samples is shown in [Figure 1]. The maximum culture positivity rate of 14.28% was observed in synovial fluid samples.
The culture positivity among males and females was found to be 57.37% and 42.63%, respectively. Age-wise distribution of patients with culture-positive sterile body fluids along with culture positivity rate is shown in [Table 3]. Department-wise distribution of the isolates is depicted in [Figure 2].
The distribution of various pathogenic bacterial isolates isolated from positive sterile body fluids cultures is shown in [Figure 3]. Among all bacterial isolates, GPC was 32 (16.67%) while GNB was 160 (83.33%).
AST pattern of GPC
The AST pattern of Staphylococcus aureus is depicted in [Table 4]. Among all 24 isolates of S. aureus, 16 (75%) were methicillin-resistant S. aureus (MRSA) in comparison to 8 (25 %) of methicillin-sensitive S. aureus (MSSA). All of the S. aureus isolates were found susceptible against vancomycin and linezolid.
AST results and distribution of various GNB along with ESBL enzyme producer as well as carbapenemase enzyme producer screening results are shown in [Tables 5 and 6].
DISCUSSION
Sterile body fluid infections contribute as important and significant causes of serious morbidity and mortality. These infections are among the common healthcare-associated infections (HCAIs). The condition can be life-threatening among patients in critical conditions especially those admitted to intensive care and high-dependency units of the hospitals.
| Age group | Total samples (n=1980) | Culture-positive samples (n=192) |
Culture positivity (%) |
|---|---|---|---|
| <10 years | 364 | 24 | 6.59 |
| 11–20 years | 196 | 12 | 6.12 |
| 21–30 years | 216 | 20 | 9.26 |
| 31–40 years | 216 | 48 | 22.22 |
| 41–50 years | 252 | 36 | 14.29 |
| 51–60 years | 320 | 32 | 10 |
| 61–70 years | 264 | 8 | 3.03 |
| >70 years | 152 | 12 | 7.89 |
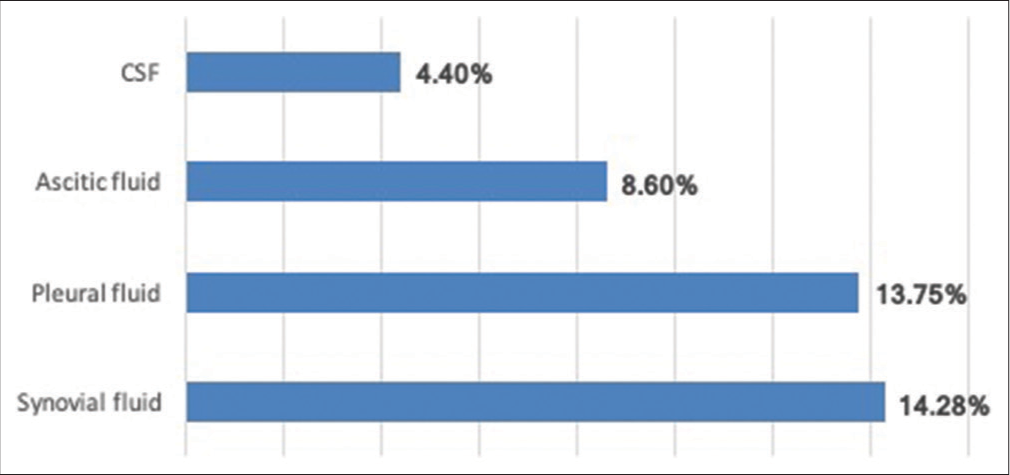
- Positivity rate of various sterile body fluid samples.

- Department-wise distribution of isolates.

- Distribution of all bacterial isolates isolated from sterile body fluids.
The culture positivity rate of sterile body fluids was found to be 9.69% in our study. Highly variable culture positivity rates were observed in various studies ranging from less than 10% to more than 50%. A higher positivity rate of 30% was observed by Sharma et al. in a study done in 2017 from north India[7] and 16.70% of positivity rate was reported by Shume et al. in 2022.[8] Around 15% of the culture positivity rate was reported by Sharma et al. in 2017, Sujatha et al. in 2015, and Vishalakshi et al. in 2016.[5,9,10] The culture positivity depends on so many factors related to clinicians, sample collection personnel, microbiologists, and factors related to the processing of the samples in the laboratory.
The clinically significant isolates were predominantly isolated from male patients (57.37%) as compared to female patients (42.63%) and the male-female ratio was found to be 1.3:1 in this study. More or less similar findings were observed from various studies in the previous years including studies done by Sharma et al. in 2018 and Teklehymanot et al. in 2017.[5,11]
Age-wise distribution of patients with culture-positive sterile body fluids revealed the higher culture positivity rate among economically productive age groups such as the age group of 31–40 years, followed by 41–50 years comprising 22.22% and 14.29%, respectively. The predominance in this age group could be due to more activities leading to more exposure to infections.
A higher proportion of the isolates (58%) were grown in the sterile body fluids samples collected from the patients admitted to the intensive care unit (ICU) as compared to other wards as ICU patients are critically ill and associated with various comorbidities and immunocompromised states hence more prone to various HCAIs.
| Antibiotic | Sensitivity of MRSA (n=16) (%) | Sensitivity of MSSA (n=8) (%) |
|---|---|---|
| Penicillin | 0 (0) | 8 (100) |
| Erythromycin | 8 (50) | 8 (100) |
| Clindamycin | 8 (50) | 4 (50) |
| Cotrimoxazole | 4 (25) | 4 (50) |
| Tetracycline | 8 (50) | 8 (100) |
| Ciprofloxacin | 8 (50) | 8 (100) |
| Moxifloxacin | 4 (25) | 4 (50) |
| Chloramphenicol | 4 (25) | 8 (100) |
| Gentamicin | 8 (50) | 8 (100) |
| Linezolid | 16 (100) | 8 (100) |
| Vancomycin | 16 (100) | 8 (100) |
| Cefoxitin | 0 (0) | 8 (100) |
AST: Antimicrobial susceptibility testing, MRSA: Methicillin-resistant Staphylococcus aureus, MSSA: Methicillin sensitive Staphylococcus aureus
| Antibiotics | Acinetobacter (n=64) | Klebsiella pneumonia (n=40) | Escherichia coli (n=24) | Pseudomonas(n=24) | Citrobacter(n=8) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | S | R | S | R | S | R | S | R | S | |
| Ampicillin | 52 | 12 | 28 | 12 | 8 | 16 | NT | NT | 4 | 4 |
| Piperacillin | 52 | 12 | 24 | 16 | 8 | 16 | 8 | 16 | 4 | 4 |
| Amoxicillin-Clavulanic acid | 44 | 20 | 20 | 20 | 12 | 12 | NT | NT | 4 | 4 |
| Ampicillin-Sulbactam | 28 | 36 | 32 | 8 | 8 | 16 | NT | NT | 0 | 8 |
| Piperacillin/Tazobactam | 48 | 16 | 12 | 28 | 8 | 16 | 8 | 16 | 0 | 8 |
| Tetracycline | 44 | 20 | 16 | 24 | 16 | 8 | NT | NT | 4 | 4 |
| Cotrimoxazole | 36 | 28 | 32 | 8 | 20 | 4 | NT | NT | 0 | 8 |
| Ciprofloxacin | 32 | 32 | 32 | 8 | 12 | 12 | 8 | 16 | 0 | 8 |
| Cefixime | 56 | 8 | 28 | 12 | 12 | 12 | NT | NT | 0 | 8 |
| Ceftazidime | 64 | 0 | 28 | 12 | 12 | 12 | 4 | 16 | 0 | 8 |
| Ceftriaxone | 64 | 0 | 28 | 12 | 12 | 12 | NT | NT | 0 | 8 |
| Aztreonam | 64 | 0 | 24 | 16 | 8 | 16 | 8 | 16 | 0 | 8 |
| Cefepime | 60 | 4 | 36 | 4 | 20 | 4 | 8 | 16 | 0 | 8 |
| Gentamicin | 24 | 40 | 28 | 12 | 4 | 20 | 8 | 16 | 0 | 8 |
| Amikacin | 16 | 48 | 24 | 16 | 4 | 20 | 8 | 16 | 0 | 8 |
| Tobramycin | 8 | 56 | 24 | 16 | 4 | 20 | 8 | 16 | 0 | 8 |
| Ertapenem | 56 | 8 | 28 | 12 | 8 | 16 | NT | NT | 0 | 8 |
| Meropenem | 56 | 8 | 28 | 12 | 8 | 16 | 8 | 16 | 0 | 8 |
| Imipenem | 52 | 12 | 24 | 16 | 8 | 16 | 8 | 16 | 4 | 4 |
| Colistin | 0 | 64 | 0 | 40 | 0 | 24 | 0 | 24 | 0 | 8 |
NT: Not tested, AST: Antimicrobial susceptibility testing, GNB: Gram-negative bacilli
| Isolated organism | ESBL screening |
Carbapenemase screening | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Acinetobacter spp. (n=64) | - | - | 52 | 12 |
| K. pneumoniae (n=40) | 28 | 12 | 28 | 12 |
| Escherichia coli (n=24) | 8 | 16 | 8 | 16 |
| Pseudomonas spp. (n=24) | - | - | 8 | 16 |
| Citrobacter spp. (n=8) | 4 | 4 | 4 | 4 |
| Total (n=160) | 40 | 32 | 100 | 60 |
ESBL: Extended-spectrum beta-lactamase
All the patients with sterile body fluid infections were having invasive devices, but the association of invasive devices with these infections could not be commented on as most of the patients were admitted to ICUs and almost all of them were having invasive devices.
In our study, there was a predominance of GNB. We observed 83.34% of GNB as compared to only 16.66% of GPC among pathogens isolated from sterile body fluids. Similarly, many studies reported predominant isolation of GNB over GPC including studies by Sharma et al., Shume et al., Sandhya et al., and Ebrahim et al. documenting 90%, 70.6%, 71%, and 74.6% of GNB in comparison to GPC isolated from sterile body fluid samples, respectively.[7,8,12,13] In a study by Bourbeau et al., no predominance of either GPCs or GNBs was observed.[14] In contrast, there are studies showing the predominance of GPC over GNB including studies by Vishalakshi et al., Çetin et al., and Pal et al.[10,15,16] This variation in the predominance of either group could be due to variation in hospital flora leading to variation among HCAIs, different hospital infection prevention and control measures followed, different sample sizes, and different geographical areas studied.[17,18]
Among the Gram-negative bacterial isolates, Acinetobacter spp. was found to be most predominant comprising 40%, followed by Klebsiella pneumoniae (25%), Escherichia coli (15%), Pseudomonas spp. (15%), and Citrobacter spp. (5%). Almost similar findings were reported in many studies including a study by Madigubba et al. with Escherichia coli at 40.10%, followed by Acinetobacter spp. 22.60%, Pseudomonas spp. 18.20%, and K. pneumoniae (14.80%).[19]
Among GPCs, S. aureus was found to be the most frequently isolated GPC comprising 75% of all GPCs.
In this study, 75% of S. aureus isolates were found to be MRSA. The reported data from the previous studies on MRSA show a wide range of variability ranging from 30% to 100% of MRSA.[8,10,15,19,20] The various factors responsible for differences in the proportion of MRSA could be geographical variations, variations in treatments followed, infection control practices and patient related factors.[4]
In this study, all S. aureus isolates were found susceptible against vancomycin and linezolid which are considered as last resort of antibiotics for the treatment of MRSA. The sensitivity pattern of S. aureus observed against other antibiotics including 50% against gentamicin, ciprofloxacin, tetracycline, clindamycin, and erythromycin each was sensitive, while 25% against chloramphenicol, moxifloxacin, and cotrimoxazole each were sensitive.
Among Gram-negative isolates, isolates 25% were ESBL producers while 62.50% were carbapenemase producers on screening while none of the isolates were resistant against colistin.
Majority of the bacterial isolates were MDROs with higher resistance against beta-lactams including cephalosporins up to third generation, aminoglycosides, and fluoroquinolones. The predominance of MDR organisms isolated from sterile body fluids was also reported in various studies including studies done by Shume et al. in 2022, Ebrahim et al. in 2020, and Tsegay et al. in 2019 documenting 76.4%, 75%, and 90% of isolates as MDROs.[8,13,21] A low proportion of 30% of isolates as MDR were documented in a study by Shrestha et al. in 2019.[20] Most of the recent studies reported majority of the pathogenic bacteria implicated in sterile body fluid infections as MDROs and our findings correlate with these recent studies.[8,13,21,22] Being a tertiary care medical teaching and referral hospital patients are referred after taking treatment at multiple hospitals in the form of antimicrobials, which can be another important reason for higher proportion of MDROs in our hospital.
The variation in the distribution of MDR organisms and their AST patterns could be due to different bacterial strains confined to that particular hospital environment, geographical variations, awareness of patients toward antibiotic usage, easy over the counter availability of antibiotics, difference in antimicrobial prescribing policies, different hospital infection control practices, and indiscriminately using antimicrobials consequently resulting in emergence and transmission of resistance against antimicrobials.[8] The higher isolation rate of MDR organisms from sterile body fluids as compared to other samples as documented by Li et al. indicates the inclusion of comparative data during the preparation of antibiogram.[22]
Limitations of the study
Anaerobic bacteria are important etiological agents associated with sterile body fluid infections. The molecular characterizations can also serve as an important modality for confirmation of bacterial identification as well as for genetic mechanisms associated with antibiotic resistance. We did not perform anaerobic culture of sterile body fluids and molecular testing for confirmation of isolates and detection of genetic mechanisms involved in antibiotic resistance due to limited resources.
CONCLUSION
From this study, we concluded that the pathogenic bacteria implicated in infections of sterile body fluids are predominantly MDR. There is huge variation in data on distribution of bacterial species isolated from sterile body fluids and their antimicrobial resistance patterns from different geographical locations and healthcare settings. Thus, data from a particular healthcare setting are important for empirical treatment in that healthcare setting.
The increasing trends of MDR and emergence of resistance against high-end antimicrobials are an alarming situation. It is the time for strict implementation of antibiotic stewardship and hospital infection control measures to prevent antimicrobial resistance.
Ethical clearance
Prior approval from the Institutional Ethics Committee was taken.
Declaration of patient consent
Patient’s consent not required as patients identity is not disclosed or compromised.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Etiology and antimicrobial resistance patterns of acute bacterial meningitis in children: A 10-year referral hospital-based study in northwest Iran. Iran Red Crescent Med J. 2014;16:e17616.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteriology of body fluids with an evaluation of enrichment technique to increase culture positivity. J Evol Med Dent Sci. 2014;3:15230-8.
- [CrossRef] [Google Scholar]
- Risk score for identifying adults with CSF pleocytosis and negative CSF Gram stain at low risk for an urgent treatable cause. J Infect. 2013;67:102-10.
- [CrossRef] [PubMed] [Google Scholar]
- Aerobic bacteriological profile and antimicrobial sensitivity pattern of bacteria isolated from sterile body fluids: A study from a tertiary care hospital in North India. Microbiol Res J Int. 2019;28:1-10.
- [CrossRef] [Google Scholar]
- Bacterial profile, their anti biogram and a light on emerging multi drug resistant organisms from sterile body fluids in a northern tertiary care hospital in India. J Bacteriol Mycol. 2018;6:249-52.
- [CrossRef] [Google Scholar]
- M100. Performance Standard for Antimicrobial Susceptibility Testing (30th ed). USA Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- [Google Scholar]
- Bacteriological profile and antimicrobial sensitivity pattern in sterile body fluids from a tertiary care hospital. J Appl Microbiol Biochem. 2017;1:1.
- [CrossRef] [Google Scholar]
- Aerobic bacterial profile and their antibiotic susceptibility patterns of sterile body fluids among patients at Hiwot Fana specialized University Hospital, Harar, Eastern Ethiopia. Infect Drug Resist. 2022;15:581-93.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteriological profile and antibiotic sensitivity pattern from various body fluids of patients attending Rama medical college hospital, Kanpur. Int J Adv Case Rep. 2015;2:119-24.
- [Google Scholar]
- A study on aerobic bacteriological profile of sterile body fluids. Int J Curr Microbiol Appl Sci. 2016;5:120-6.
- [CrossRef] [Google Scholar]
- Bacterial profile and their antimicrobial resistance patterns from body fluids at Tikur Anbesa Specialized Hopital, Addis Ababa, Ethiopia. Biol Med. 2017;9:408.
- [CrossRef] [Google Scholar]
- A study of bacteriological profile of sterile body fluids in a tertiary care hospital. Int J Sci Res. 2019;8:41-5.
- [Google Scholar]
- Bacterial profile and antimicrobial susceptibility pattern of isolates recovered from sterile body fluids referred to the national reference laboratory. Lancet Planet Health. 2020;4:e379-80.
- [CrossRef] [PubMed] [Google Scholar]
- Use of the BacT/Alert blood culture system for culture of sterile body fluids other than blood. J Clin Microbiol. 1998;36:3273-7.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of the BACTEC blood culture system versus conventional methods for culture of normally sterile body fluids. Adv Ther. 2007;24:1271-7.
- [CrossRef] [PubMed] [Google Scholar]
- Optimum time to detection of bacteria and yeast species with BACTEC 9120 culture system from blood and sterile body fluids. J Lab Physicians. 2009;1:69-72.
- [CrossRef] [PubMed] [Google Scholar]
- Guidelines for prevention of hospital acquired infections. Indian J Crit Care Med. 2014;18:149-63.
- [CrossRef] [PubMed] [Google Scholar]
- Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804-13.
- [CrossRef] [PubMed] [Google Scholar]
- Bacteriological profile and antimicrobial susceptibility pattern in sterile body fluid specimens from a tertiary care hospital, South India. J Curr Res Sci Med. 2020;6:96-101.
- [Google Scholar]
- Bacteriological profile and antimicrobial susceptibility pattern among isolates obtained from body fluids. J Nepal Health Res Counc. 2019;17:173-7.
- [CrossRef] [PubMed] [Google Scholar]
- Bacterial isolates and drug susceptibility pattern of sterile body fluids from tertiary hospital, Northern Ethiopia: A four-year retrospective study. J Pathog. 2019;2019:5456067.
- [CrossRef] [PubMed] [Google Scholar]
- Study on the detection and infection distribution of multidrug-resistant organisms in different specimens. Infect Drug Resist. 2022;15:5945-52.
- [CrossRef] [PubMed] [Google Scholar]






