Translate this page into:
Usefulness of truenat-derived DNA from extrapulmonary specimens in direct detection of drug resistant tuberculosis by line probe assay
*Corresponding author: Sarika Jain Agrawal, Laboratory Division, National Tuberculosis Institute, Bengaluru, Karnataka, India. sarika.jainagrawal@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Jain Agrawal S, Mamatha V, Somashekar N. Usefulness of truenat-derived DNA from extrapulmonary specimens in direct detection of drug resistant tuberculosis by line probe assay. Indian J Med Sci 2023;75:168-73.
Abstract
Objectives:
Isoniazid (INH) and second-line drug resistance (DR) detection through line probe assay (LPA) takes long in extrapulmonary (EP) specimens because culture growth needs to be obtained to perform deoxyribonucleic acid (DNA) extraction due to the paucibacillary nature of these specimens. Knowing the DR pattern at the earliest is key to success of the treatment. Delay in appropriate tuberculosis (TB) treatment in EP TB patients runs the risk of DR amplification, significant disease damage, and patient loss to follow-up. Here, LPA was attempted on truenat-derived DNA elute from EP specimens, which, in routine, is discarded after the truenat test, to determine drug sensitivity test (DST) for INH and, where necessary, for second-line drugs (Fluoroquinolones, Kanamycin, amikacin, and capreomycin).
Material and Methods:
Truenat, acid-fast bacilli culture, and fluorescent microscopy were performed on all EP samples that were received at the laboratory during June–September 2022. DNA elute that was left over from 59 truenat Mycobacterium tuberculosis (MTB) positive EP samples were subjected to Genotype MTBDR plus Ver 2.0 assay.
Results:
MTBDR plus assay (DNA elute) detected MTB and rifampicin (RIF) and INH DST in 47 samples (79.6%) having truenat MTB count of 7.8 × 102 colony-forming unit/milliliter and above. It also detected RIF DST in 65.2% truenat RIF indeterminate samples and DST for both RIF and INH in 60% of culture negative EP specimens. DST results by LPA (DNA elute) completely concorded with standard indirect LPA (on 21 culture isolates from smear-negative specimens). The MTBDRsl yield was however relatively low (11.1%), although second line LPA (SLLPA) was performed only on 9 first-line DR samples.
Conclusions:
Left-over truenat-derived DNA elute is a significant sample by-product that can significantly speed up and increase the yield of determination of MTB DST in EP samples for RIF and INH, the most critical drugs for TB treatment.
Keywords
Extrapulmonary tuberculosis
Truenat
Line probe assay
MTB DR plus
DNA elute
INTRODUCTION
The rapid nucleic acid amplification test (NAAT) Truenat™ (Molbio Diagnostics, Goa, India) allows for the quantitative detection of Mycobacterium tuberculosis (MTB) and resistance to rifampicin (RIF) in direct sputum and extrapulmonary (EP) specimens.[1] The system uses the Trueprep® AUTO v2 Universal Cartridge-based Sample Prep Device and the Truelab® Real Time mini polymerase chain reaction (PCR) Analyzer, respectively, to automate the extraction and purification of deoxyribonucleic acid (DNA) and real-time PCR. Rapid NAAT has been suggested under the National Tuberculosis Elimination Program of India (NTEP) as the initial diagnostic test for the early diagnosis of extrapulmonary (EP) tuberculosis (EPTB).[2]NAATs (GeneXpert or Truenat) offer rapid and direct RIF resistance identification in clinical specimens; however, INH and second-line drug resistance (DR) detection takes longer in EP specimens because the bulk of these specimens is paucibacillary in nature.
Due to the suboptimal sensitivity of MTBDRplus and MTBDRsl assays on smear-negative material, MTB culture growth must be obtained before moving on to DNA extraction for the first-line and second-line drugs line probe assays (LPA).[3,4] This can take a month time or even longer.
In addition, after 6–8 weeks of incubation, smear-negative EP specimens may turn out to be culture-negative or may even be lost because culture is prone to contamination. To retry culture, repeat patient sampling would need to be undertaken, which is difficult in EPTB patients. In these circumstances, the patient’s tuberculosis (TB) treatment will continue based only on the NAAT results of RIF drug resistance testing (DRT). Even RIF DRT information is inaccessible if RIF resistance is indeterminate in NAAT. Delaying the start of drug sensitivity test (DST)-based appropriate TB therapy in these patients runs the risk of DR amplification, significant disease damage, and patient loss to follow-up.
The prevalence of INH mono resistance is 6–8% in EP TB cases according to data from studies conducted in India and around the world,[5] which if not treated properly may drive further RIF resistance. RIF mono-resistance is 3–7%, and multi-DR (MDR) prevalence in EPTB varies from 3% to 13.5%.[6-8] MDR can frequently cause treatment failure and fatalities.
In a study conducted in southern India to determine the proportion of patient samples that underwent the complete DR-TB diagnostic algorithm recommended under the national TB program, it was found that 27% of patient samples did not reach the TB reference laboratories for additional testing after gene Xpert. Of those samples that did reach the laboratories, 7% did not complete the DR-TB diagnostic algorithm. Non-completion of the algorithm was more common with extrapulmonary, sputum smear-negative, and RIF resistant-TB samples. Due to culture negativity in smear-negative samples or problems with specimen transportation to affiliated TB laboratories, LPA could not be performed on these samples.[9]
An alternate molecular-based strategy is hence urgently needed in the TB care cascade to quickly detect DR to first- and second-line anti-TB drugs in direct samples and offer evidence-based TB treatment to patients because treatment for EPTB is likely to last longer depending on the site, severity, and DR.
In this report, we outline our attempt to perform LPA on truenat-derived DNA elute from EP specimens to determine DST for INH and, where necessary, for the second-line drug. We were prompted to attempt the present work to address the concerns of clinicians in determining at the earliest whether to prescribe shorter or longer MDR treatment regimens in patients with RIF resistance or indeterminate resistance in NAAT and also for patients who had an extensive disease or were at risk of DR. Therefore, the current work was not intended to prospectively evaluate that the utility of leftover DNA elutes from a truenat test for LPA testing rather is presented here as our clinical observation. Since LPA was performed on leftover DNA which is discarded in routine after truenat test, patient consent was not required and ethics approval was not needed. Further, LPA (DNA elute) results were not formally released. The results were conveyed to clinicians for patient management only for standard LPA tests performed on smear-positive samples or culture isolates when the results were available.
MATERIALS AND METHODS
All EP samples received at our laboratory under the NTEP are subjected to truenat MTB plus test, liquid culture (MGIT 960 automated system), and fluorescence microscopy for diagnosis of TB. Truenat,[10,11] LPA,[12,13] and liquid culture[14] are performed as per the manufacturer’s instructions. Routinely, direct LPA is performed on DNA extracted from smear-positive specimens while indirect LPA is on culture growth from smear-negative specimens after the specimen has been decontaminated and concentrated using the NALC-NaOH procedure.
DNA elute left over from truenat MTB positive EP samples that were received at our NRL during June–September 2022 was subjected to LPA. The workflow for EP specimen processing is depicted in [Figure 1]. LPA (DNA elute) was initially conducted on two of the EP specimens at the clinician’s request, and the results were compared with those from standard indirect LPA on culture isolates. LPA on DNA elute of additional EP samples was tried only after both LPAs showed complete concordance.
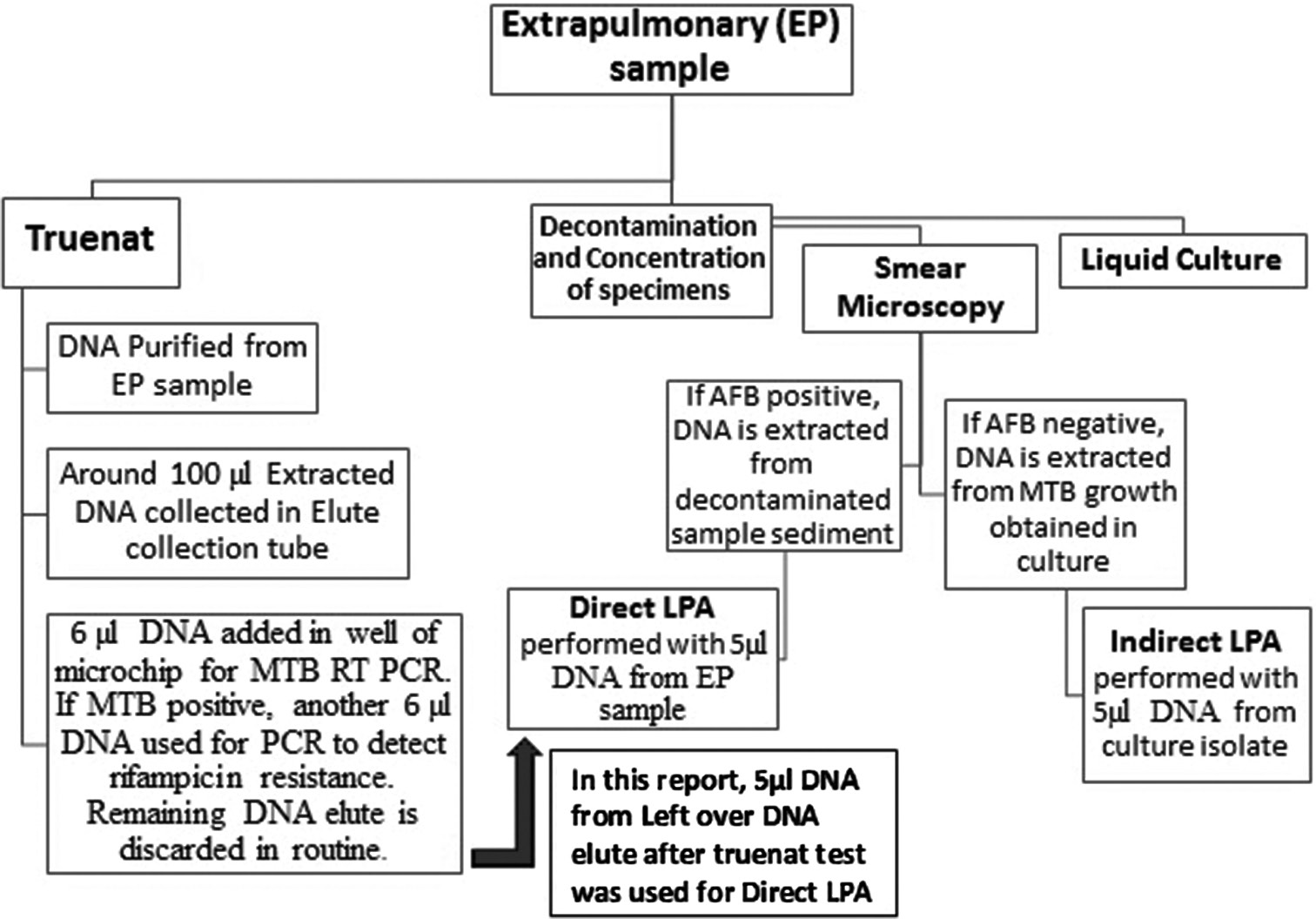
- Flowchart outlining the extrapulmonary sample workflow at our tuberculosis lab.
Briefly, MTBDRplus version 2 and MTBDRsl version 2 LPAs were performed using 5 µL of DNA volume from 70 µL to 88 µL of leftover DNA elute after the truenat test was finished without any additional purification steps or processing of DNA elutes. All EP samples were subjected to MTBDRplus, whereas MTBDRsl was only carried out on samples that were RIF or INH resistant in MTBDRplus assay. DNA elutes were stored at 4°C after the truenat test until LPA on these elutes was performed.
RESULTS
Truenat test was positive for MTB in 59 EP samples received during June–September 2022.
The EP samples comprised peritoneal fluid, pleural fluid, urine, lymph node aspirate, pus from lymph node abscess, lymph node biopsy, spinal tissue biopsy, and various tissue biopsies. Patients’ ages ranged from 12 to 79 years old (26 were male, 33 were female).
On fluorescent microscopy, smears from only two samples were positive whereas 57 samples were negative for acid-fast bacilli.
First-line LPA-FLLPA (MTBDR plus) was attempted on leftover DNA elute from these 59 truenat MTB complex positive extrapulmonary samples in the present report.
MTBDR plus assay on truenat DNA elutes
Using the DNA elute-based MTBDR plus assay, valid and clear MTB and RIF DST were obtained in 48 out of 59 samples (81.3%), and INH DST results in 47 samples (79.6%). There were no background contamination bands visible on any of the 48 first-line LPA hybridization blot strips. Overall, 47 samples showed valid MTB and DST results for both drugs and 12 (20.3%) samples didn’t show complete results; 11 samples lacked the TUB band (M.tuberculosis) for MTB and 1 sample lacked the gene loci for RIF and/or INH drugs [Table 1].
| Result | MTB DRplus (on 59 truenat DNA elute) | MTBDRsl (on 9 MTBDR plus resistant truenat DNA elute#) |
|---|---|---|
| MTB detected | 48/59 (81.3%) | 1/9#(11.1%) |
| MTB Not detected | 11/59 (18.6%) | 8/9#(88.9%) |
| Valid DST results for all Drugs in | 47/59 (79.6%) | 1/9 (11.1%) |
| MTB positive samples | ||
| Any drug resistance detected | 9#/47 (19.1%) Rifampicin- 3/48 (6.3%) INH- 4/47 (8.5%) MDR- 3/47 (6.4%) |
1/1 (100%) Fluoroquinolone resistant- 1/1 |
#Drug resistant samples in MTB DRplus were subjected to MTBDRsl.
MTB: Mycobacterium tuberculosis, DR: Drug resistance, DNA: Deoxyribonucleic acid, DST: Drug sensitivity test, INH: Isoniazid, MDR: multi-DR
Drug resistance (DR)
In 32 samples where RIF results were available by both truenat and LPA (DNA elute), 100% RIF concordance was seen. Truenat detected RIF resistance in 2 out of 59 samples (3.4%). Both samples were also found RIF resistant (and susceptible to INH) on MTBDR plus assay (DNA elute). Three samples that were truly RIF sensitive were INH resistant by LPA.
[Table 2] depicts comparison between rifampicin DST in 48 samples by Truenat and LPA (from DNA elute), their MTB count, INH DST and status of DST for second-line drugs (in 9 samples). RIF findings in Truenat were indeterminate in 23 of the 59 samples (38.9%). Importantly, valid RIF results were obtained by LPA (DNA elute) in 15 of these 23 (65.2%) truenat RIF indeterminate samples; three of these samples were MDR (RIF and high level INH resistant); and one sample was INH mono-resistant (low level).
| Rifampicin DST | MTBDR plus on DNA Elute (n=48) | Truenat MTB count range (CFU/ml) |
MTBDR sl (n=9) | |||
|---|---|---|---|---|---|---|
| Truenat | MTBDR plus on DNA Elute (n=48) | RIF Comparison between truenat & LPA on DNA elute | INH DST | Fluoroquinolone DST | Second line Injectables DST |
|
| Resistant (n=2) | Resistant (2) | 100% Concordance | Susceptible (2) | 4.4×103 & 5.3×103 |
Resistant (1/2) | Susceptible (1/2) |
| No result (1/2) | ||||||
| Indeterminate (n=23) |
Resistant (3) | Rif results obtained in LPA for 15 (65.2%) truenat RIF indeterminates |
High level Resistant (3) |
4.3×101-1.9×104 | No result (1/3) Background contamination (2/3) |
|
| Susceptible (12) | Susceptible (11) Low level Resistant (1) |
ND (11) No result (1/1) |
||||
| MTB not detected (8) |
8 (34.8%) RIF not determined |
- | 1.5×102-1.1×103 | ND (8) | ||
| Susceptible (n=34) | Susceptible (30) | 98.7% Concordance | Susceptible (27) High level Resistant (3) |
3.9×102-6.9×105 | ND (27) Background contamination (3/3) |
|
| MTB not detected (4) | 1.3% RIF not determined | - | 3.9×101-1.1×103 | |||
No result: MTB & DST bands were absent; DST: Drug susceptibility test ; ND: Not performed as no resistance detected in MTBDR plus. MTB: Mycobacterium tuberculosis, DR: Drug resistance, DNA: Deoxyribonucleic acid, INH: Isoniazid, MDR: multi-DR, EP: Extrapulmonary, CFU: Colony-forming unit, RIF: Rifampicin, LPA: line probe assay
MTBDR sl assay on truenat DNA elute
However, only one sample (11.1%) from these nine DR samples, using the MTBDRsl assay on DNA elutes produced valid MTB and DST results and was identified as fluoroquinolone-resistant. The other eight samples did not produce a TUB band for MTB or had background contamination bands and were uninterpretable.
MTB count in truenat
The mycobacterial load in the samples that were truenat MTB positive ranged from 3.9 × 101 to 6.9 × 105 colony-forming unit (CFU)/milliliter (mL). In the majority of our samples with valid FLLPA DST results (43/47), MTB count was equivalent to or more than 7.8 × 102 CFU/mL with the exception of four samples with truenat MTB count of 4.3 × 101 to 5.4 × 102. [Table 3] shows truenat MTB count and RIF results along with culture positivity details of 12 samples where either TUB band or RIF/INH DST were not available by LPA (DNA elute). The MTB count ranged in these samples from 3.9 × 101 to 7.6 × 102 with the exception of three samples (7.8 × 102 in 1 sample, and 1.1 × 103 CFU per ml in 2 samples).
| Truenat | CBNAAT done in truenat RIF indeterminate samples |
MTBDR plus (DNA Elute) | MTB Culture |
Specimen type | ||||
|---|---|---|---|---|---|---|---|---|
| Rifampicin DST | MTB count (CFU/ml) |
MTB status |
RIF DST | MTB | RIF DST | INH DST | ||
| Indeterminate (8) | 7.6×102 | Not detected | NA | TUB absent |
gene Locus absent | gene Locus absent | Positive | FNA Lymph Node |
| 7.6×102 | Very Low | Indetermina te | TUB absent |
gene Locus absent | gene Locus absent | Positive | FNA Lymph Node | |
| 6.9×102 | Low | Sensitive | TUB absent |
gene Loci absent | Sensitive | Positive | Tissue biopsy | |
| 5.1×102 | Low | Sensitive | TUB absent |
gene Locus absent | gene Locus absent | Negative | FNA Lymph Node |
|
| 7.6×102 | Very Low |
Sensitive | TUB absent |
gene Locus absent | gene Locus absent | Negative | FNA Lymph Node |
|
| 1.1×103 | Very Low |
Sensitive | TUB absent |
gene Locus absent | gene Locus absent | Negative | Ascitic Fluid | |
| 7.8×102 | Very Low |
Sensitive | TUB absent |
gene Locus absent | gene Locus absent | Negative | Pleural Fluid | |
| 1.5×102 | Low | Resistant | TUB present |
Resistant | gene Locus absent | Negative | FNA Lymph Node |
|
| Susceptible (4) |
3.9×101 | Not done |
TUB absent |
gene Locus absent | gene Locus absent | Negative | Tissue biopsy | |
| 2.7×102 | Not done |
TUB absent |
gene Locus absent | gene Locus absent | Positive | Tissue biopsy | ||
| 1.1×103 | Very Low | Sensitive | TUB absent |
gene Locus absent | gene Locus absent | Negative | Cervical Lymph Node biopsy |
|
| 2.6×102 | Not done | TUB absent |
gene Locus absent | gene Locus absent | Negative | Cervical Lymph Node biopsy | ||
Footnote: TUB band: MTB; gene locus: locus for target gene for drugs-INH, Rifampicin; DST: Drug susceptibility test; MTB: Mycobacterium tuberculosis, DR: Drug resistance, DNA: Deoxyribonucleic acid, CFU: Colony-forming unit, RIF: Rifampicin, INH: Isoniazid, FNA: Fine needle aspirate
Liquid culture and indirect LPA from culture isolates
The MGIT 960 automated liquid culture method detected MTB growth in 21 out of 59 samples (35.6%). Later, results of the routine indirect MTBDR plus results from 21 MTB culture isolates concorded with that of LPA results from DNA elutes for these samples. Comparing MTBDR plus (DNA elute) to truenat and indirect LPA from liquid culture isolate, there were no false resistant or sensitive DST results.
Indirect MTBDRsl from culture isolates of first-line resistant samples produced valid results in three samples as opposed to just 1 sample when MTBDRsl from DNA elutes was performed. FQ resistant result was found for the same sample in both MTBDRsl assays.
DISCUSSION
In this report, we discuss our clinical experience with conducting LPA on leftover DNA that was extracted from truenat MTB-positive clinical EP specimens.
In a study by Venter et al, MTBDRplus and MTBDRsl tests were run using cartridge extract (CE) from used Xpert TB-positive cartridges. On CE from clinical sputum specimens and culture isolates, they found high frequencies of false-RIF resistance and indeterminate resistance results with MTBDRplus assay. However, MTBDRsl demonstrated low indeterminate rates and good accuracy for both drug-sensitive-TB and DR-TB specimens when the Xpert semi-quantitation category was at least “medium” or CT 24 (equivalent to 103 CFU/mL).[15]
In contrast to the latter study, we observed good DST accuracy with MTBDRplus assay on truenat positive DNA elutes when the MTB count in truenat was at least 7.8 × 102 CFU/mL.
The SLLPA yield was however relatively low (11.1%) in this report although SLLPA was performed only on 9 first-line DR samples; hence, it is difficult to draw conclusions about its performance on DNA elutes.
It is noteworthy that out of 59 samples, only 21 yielded MTB-positive cultures, while the remaining 38 samples (64.4%) were lost because either the cultures were contaminated (7 samples) or growth negative (31 samples). In contrast, LPA (DNA elute) findings were obtained for 23 of these 38 culture-negative samples (60.5%), for which otherwise DST for RIF in truenat indeterminate samples and INH DST could not have been determined.
Further, time to positivity for MTB growth in liquid culture was 18–35 days, and the turnaround time (TAT) for indirect LPA from MTB culture isolates was 21–38 days long. In contrast, TAT for LPA results from DNA elutes after truenat testing was only 1–5 days.
CONCLUSION
Our results show that truenat-derived DNA elute can be a significant sample by-product that can (a) aid in detecting LPA DST in 60% of culture-negative EP specimens with MTB count of 7.8 × 102 CFU/mL or more and (b) significantly shorten the time for determination of MTB DST for the most critical drugs for TB treatment – RIF and isoniazid (INH)-in direct EP samples (within 1–5 days). Appropriate TB treatment may be otherwise delayed in these patients by almost a month or more by routine indirect LPA on EP samples. (c) In addition, it can offer potentially reliable RIF DRT results in 65% of samples that were truenat RIF indeterminate.
As far as we are aware, this is the first report of LPA performed on truenat-extracted DNA from direct EP specimens, which otherwise would have been discarded following the truenat test.
In programmatic circumstances, prompt delivery of specimens following NAAT testing to the affiliated TB laboratories for LPA, culture, and DST is crucial for the accuracy and timeliness of the diagnosis. The field facts, as seen in the study,[4] and our everyday experience, however, indicate that transit delays of specimens or their failure to arrive at the laboratory are not unusual. (d) Since DNA is more durable than bacilli during transportation and storage, transporting DNA elute with at least MTB count 7.8 × 102 CFU/mL from the truenat location to the linked reference laboratory together with the specimen material may help answer the concerns over contamination or loss of viability of mycobacterium in culture.
The rapidity and accuracy of LPA DST results obtained on truenat-derived DNA from our EP samples, completely concordant with that of LPA DST on culture isolates, are very encouraging. As these findings can have a significant positive bearing on patient management, larger and more comprehensive studies are warranted in multiple settings and under programmatic field conditions to test the expected outcomes in comparison with the reference standard and to further understand the effect of storage and transportation of DNA elute on DST results; the relationship between truenat mycobacterial load and DST outcomes in LPA (DNA elute). If found promising, the practical viability of incorporating DNA elutes for LPA in the national TB program’s diagnostic algorithm may be considered.
Declaration of patient consent
Patient’s consent not required as patients identity is not disclosed or compromised.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Rapid Communication: Molecular Assays as Initial Tests for the Diagnosis of Tuberculosis and Rifampicin Resistance. 2020. Geneva: World Health Organization; Available from: https://www.who.int/publications/i/item/9789240000339 [Last accessed on 2023 Jun 26]
- [Google Scholar]
- Guidelines for Programmatic Management of Drug Resistant TB in India. 2021. Available from: https://tbcindia.gov.in/showfile.php?lid=3625 [Last accessed on 2023 Jun 26]
- [Google Scholar]
- The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Sci Rep. 2016;6:17850.
- [CrossRef] [PubMed] [Google Scholar]
- The diagnostic accuracy of the GenoType® MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev. 2014;10:CD010705.
- [CrossRef] [Google Scholar]
- Drug resistance and its risk factors among extrapulmonary tuberculosis in Ethiopia: A systematic review and meta-analysis. PLoS One. 2021;16:e0258295.
- [CrossRef] [PubMed] [Google Scholar]
- Drug resistance among extrapulmonary TB patients: Six years' experience from a supranational reference laboratory. Indian J Med Res. 2015;142:568-74.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug-resistant pulmonary & extrapulmonary tuberculosis: A 13 years retrospective hospital-based analysis. Indian J Med Res. 2015;142:575-82.
- [CrossRef] [PubMed] [Google Scholar]
- Rising trend of drug resistance among extra pulmonary TB in Northern India. Tuberculosis. 2018;52:PA3681.
- [CrossRef] [Google Scholar]
- Implementation of the new integrated algorithm for diagnosis of drug-resistant tuberculosis in Karnataka State, India: How well are we doing? PLoS One. 2021;16:e0244785.
- [CrossRef] [PubMed] [Google Scholar]
- Truenat™ MTB Package Insert. 2020. Available from: https://www.molbiodiagnostics.com/uploads/product_download/20211215.174722~TruenatMTB-packinsert-Version-06.pdf [Last accessed on 2023 Feb 28]
- [Google Scholar]
- Truenat™ MTB-RIF Dx. 2020. Available from: https://www.molbiodiagnostics.com/uploads/product_download/20220502.105606~Truenat-MTBRIF-Dx-packinsert-V05.pdf [Last accessed on 2023 Feb 28]
- [Google Scholar]
- MGIT Procedure Manual for BACTEC™ MGIT™ 960 TB System. 2006 Available from: https://www.finddx.org/wp-content/uploads/2023/02/20061101_rep_mgit_manual_FV_EN.pdf [Last accessed on 2023 Jun 26]
- [Google Scholar]
- Mycobacterial genomic DNA from used Xpert MTB/RIF cartridges can be utilised for accurate second-line genotypic drug susceptibility testing and spoligotyping. Sci Rep. 2017;7:14854.
- [CrossRef] [PubMed] [Google Scholar]






